Towards Scrutinizing the Impact of the piRNA Pathway in Human Male Infertility
Infertility represents a huge psychological burden for affected men. Receiving a causative diagnosis not only aids to cope with their situation but also helps to estimate chances for medically assisted reproduction (MAR). In a collaborative effort, researchers from the University of Münster, Gießen, Edinburgh, Newcastle, Strasbourg, Barcelona and Nijmegen demonstrate that genetic variation leading to the disruption of the piRNA pathway machinery can be linked to male infertility.
Full details of the findings were published this week in Nature Communications in a paper titled, “Inherited defects of piRNA biogenesis cause transposon de-repression, impaired spermatogenesis, and human male infertility.”
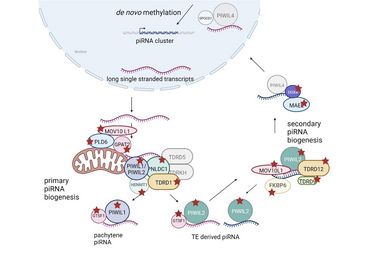
piRNAs represent a subgroup of small RNA molecules predominantly expressed in the mammalian testis. They bind to certain Argonaut proteins, known as PIWI proteins, and contribute to the repression of transposons through de novo methylation of genomic DNA and post-transcriptional silencing and are, thus, essential for maintaining genome integrity. In addition, in the adult testis, they also contribute to the regulation of gene expression. While the function of piRNAs has been well-characterised in mice, their role in human germ cell development is less understood.
The authors identified a significant association between homozygous loss-of-function variants found in exome/genome data of men with spermatogenic failure and genes involved in “piRNA processing”. In a subsequent screening approach of genomic data from four independent cohorts of infertile men, 39 men carrying high-impact biallelic variants in 14 different piRNA-pathway related genes were identified.
Following on from this, the authors found that disturbed piRNA biogenesis in men is associated with a de-repression of transposable elements. By characterising the reproductive and testicular phenotypes of affected men, they show concordant gene-specific findings that highlight important differences between humans and mice, indicating that findings from mouse models are not generally valid for all mammals.
In essence, this study marks a substantial advancement in the field of reproductive genetics by significantly expanding the number of candidate genes linked to male infertility and establishing a connection between genetically disrupted piRNA biogenesis and male infertility. The success of this work is rooted in both the collaborative collection of exome sequencing data of infertile men and in-depth clinical characterisation of the associated reproductive phenotypes as well as specific laboratory expertise and expertise on the piRNA biogenesis field.